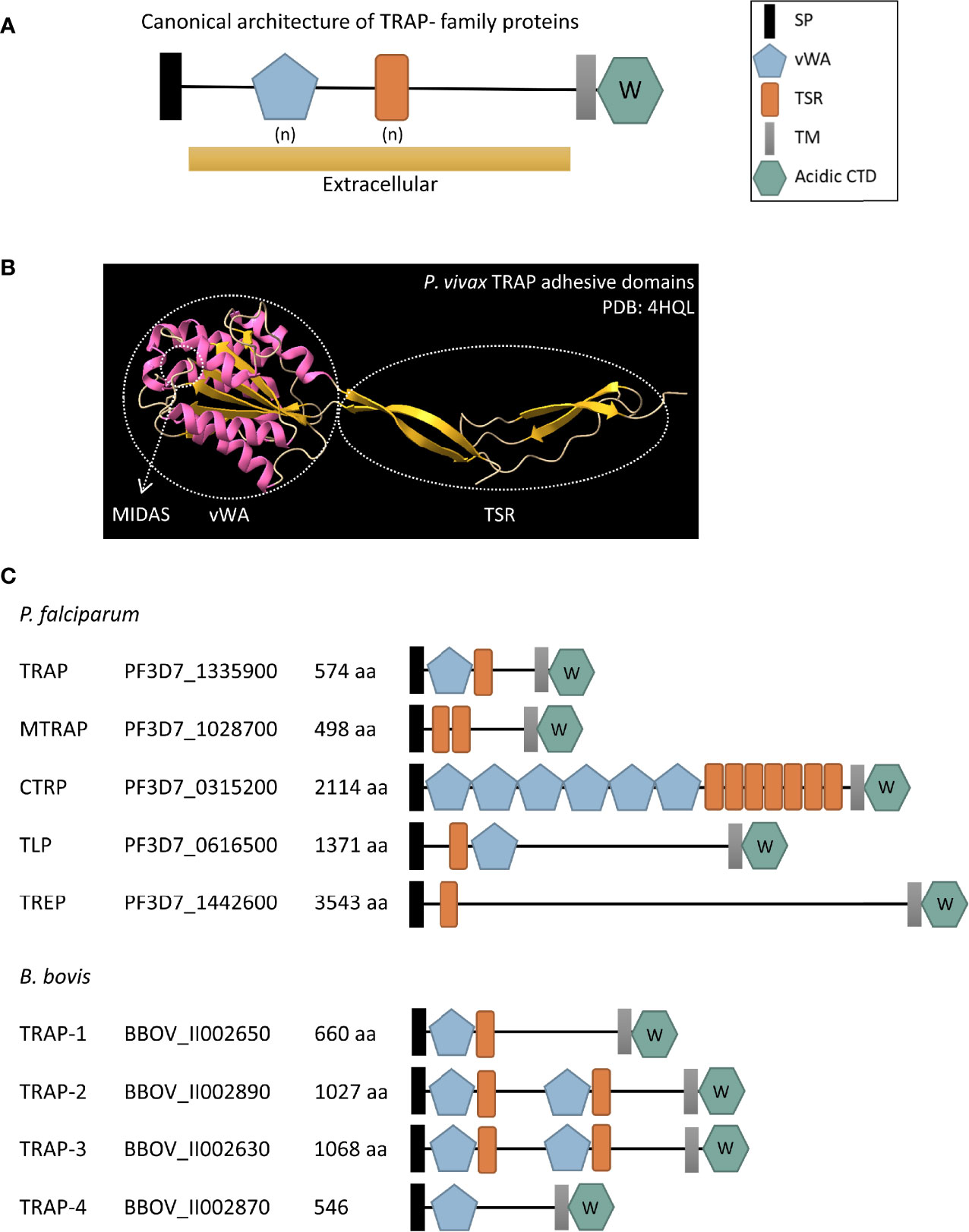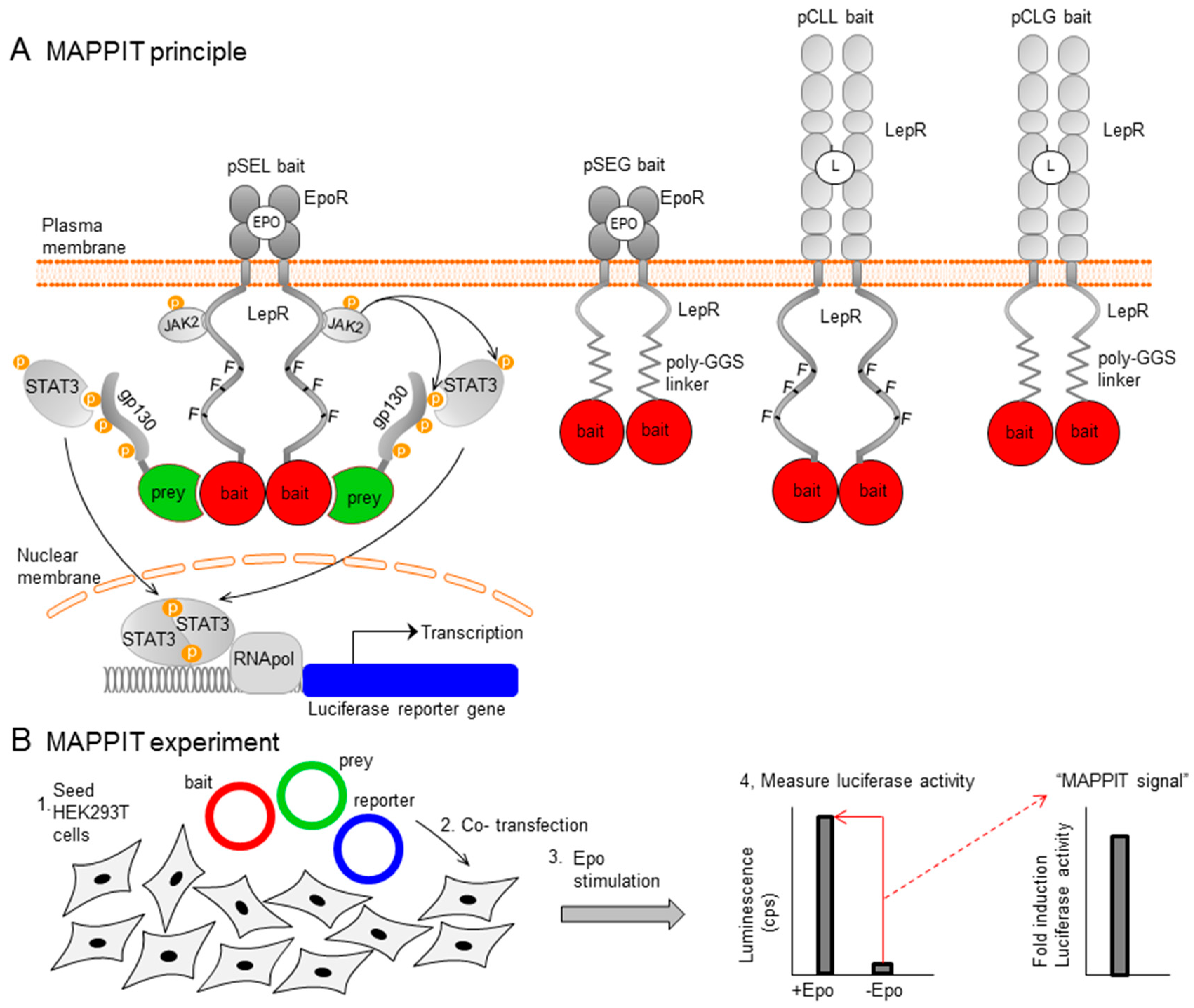
IJMS | Free Full-Text | Straightforward Protein-Protein Interaction Interface Mapping via Random Mutagenesis and Mammalian Protein Protein Interaction Trap (MAPPIT)

Deep Bottom-up Proteomics Enabled by the Integration of Liquid-Phase Ion Trap | Analytical Chemistry

Molecular basis of the TRAP complex function in ER protein biogenesis | Nature Structural & Molecular Biology

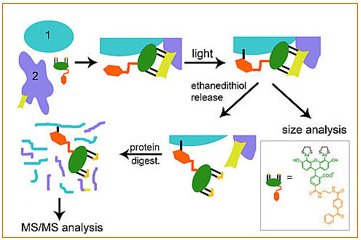
A molecular trap inside microtubules probes luminal access by soluble proteins | Nature Chemical Biology

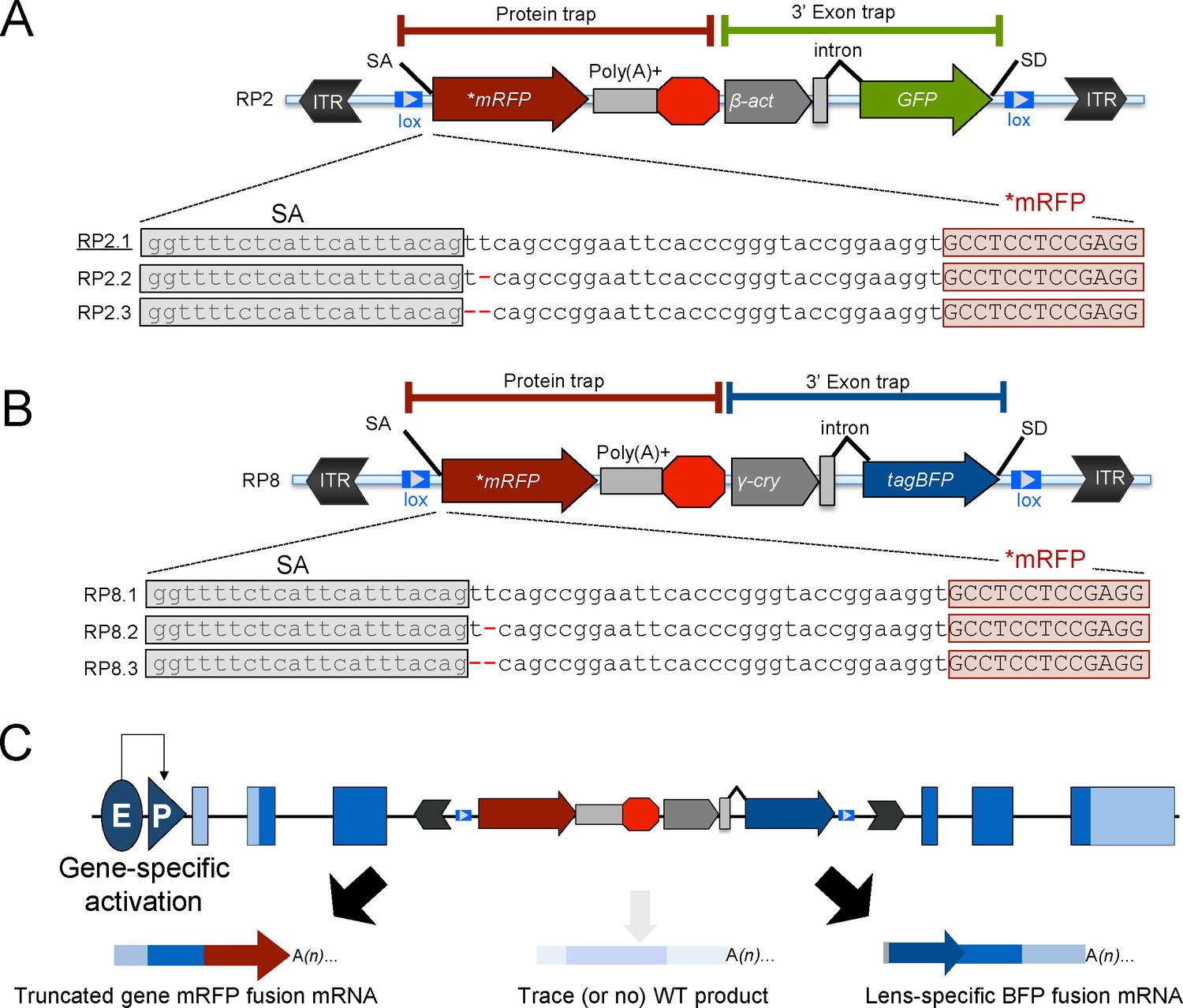
A split green fluorescent protein system to enhance spatial and temporal sensitivity of translating ribosome affinity purification - Dinkeloo - 2022 - The Plant Journal - Wiley Online Library

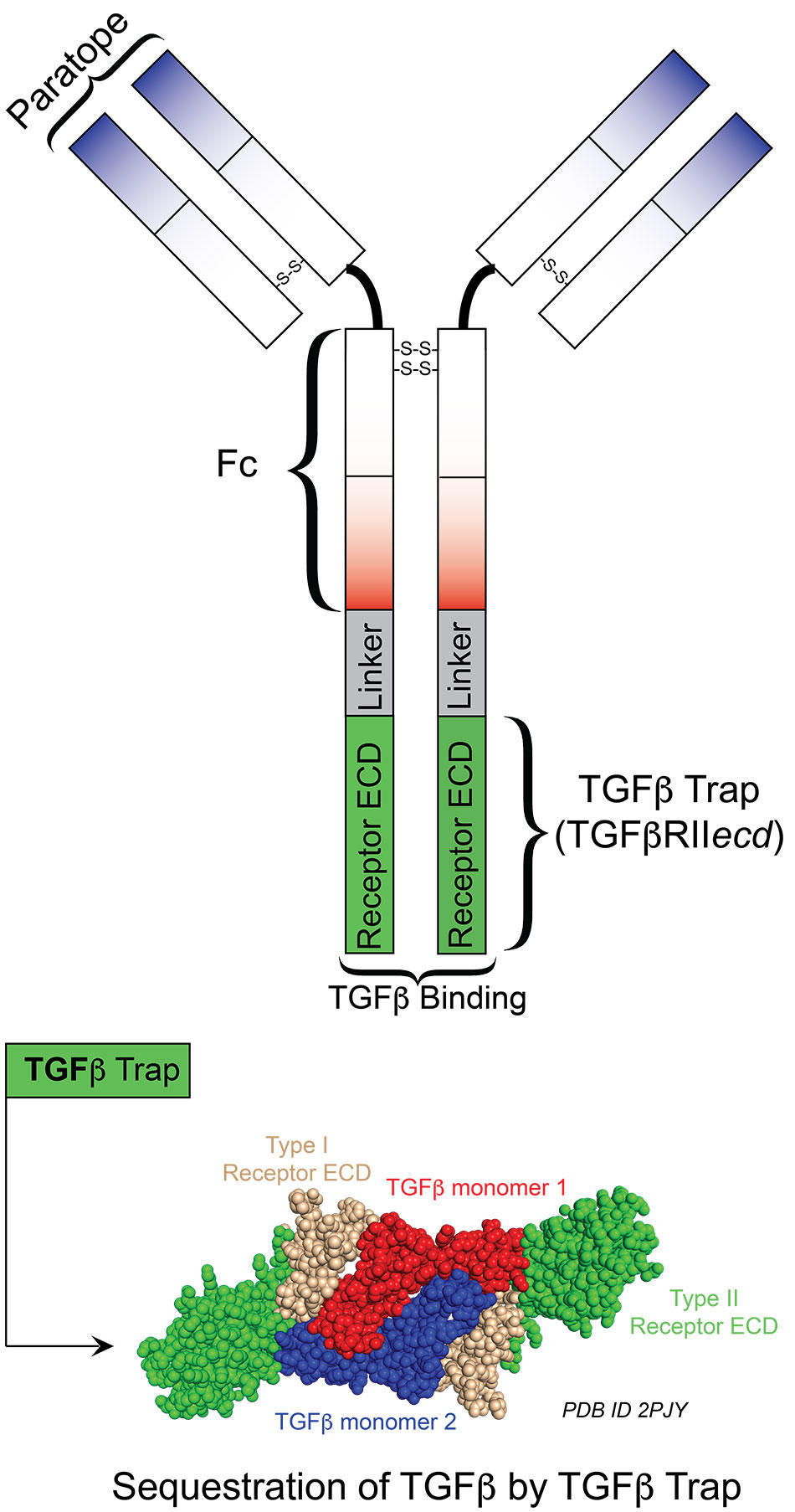
JCI Insight - TGF-β1 protein trap AVID200 beneficially affects hematopoiesis and bone marrow fibrosis in myelofibrosis
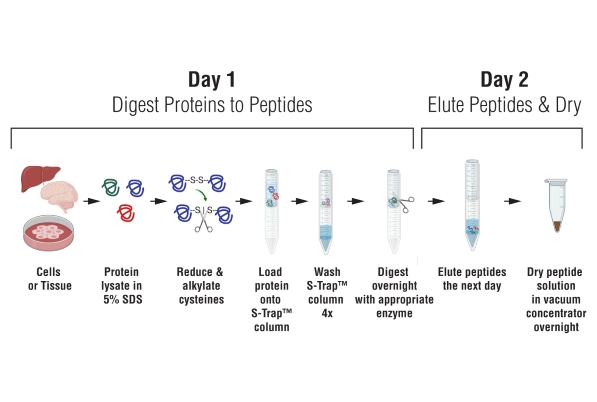
PSR Protocol for S-Trap micro spin column digestion Ref: https://www.protifi.com/resources/ Reagents Sodium DodecylSulfate (SDS

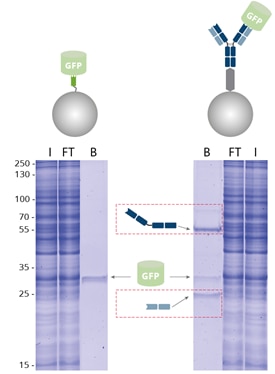
Proteintech ChromoTek GFP-Trap™ Magnetic Particles M-270 20 Reactions Proteintech ChromoTek GFP-Trap™ Magnetic Particles M-270 | Fisher Scientific

Secretion of bispecific protein of anti-PD-1 fused with TGF-β trap enhances antitumor efficacy of CAR-T cell therapy: Molecular Therapy - Oncolytics

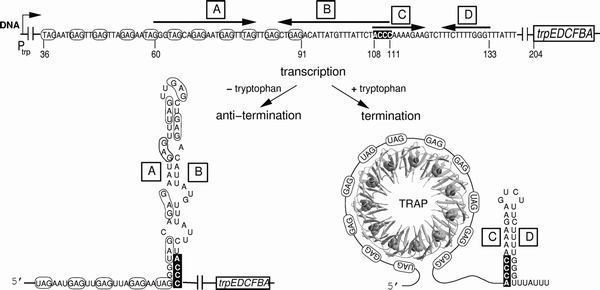
Crystal structure of Bacillus subtilis anti-TRAP protein, an antagonist of TRAP/RNA interaction | PNAS
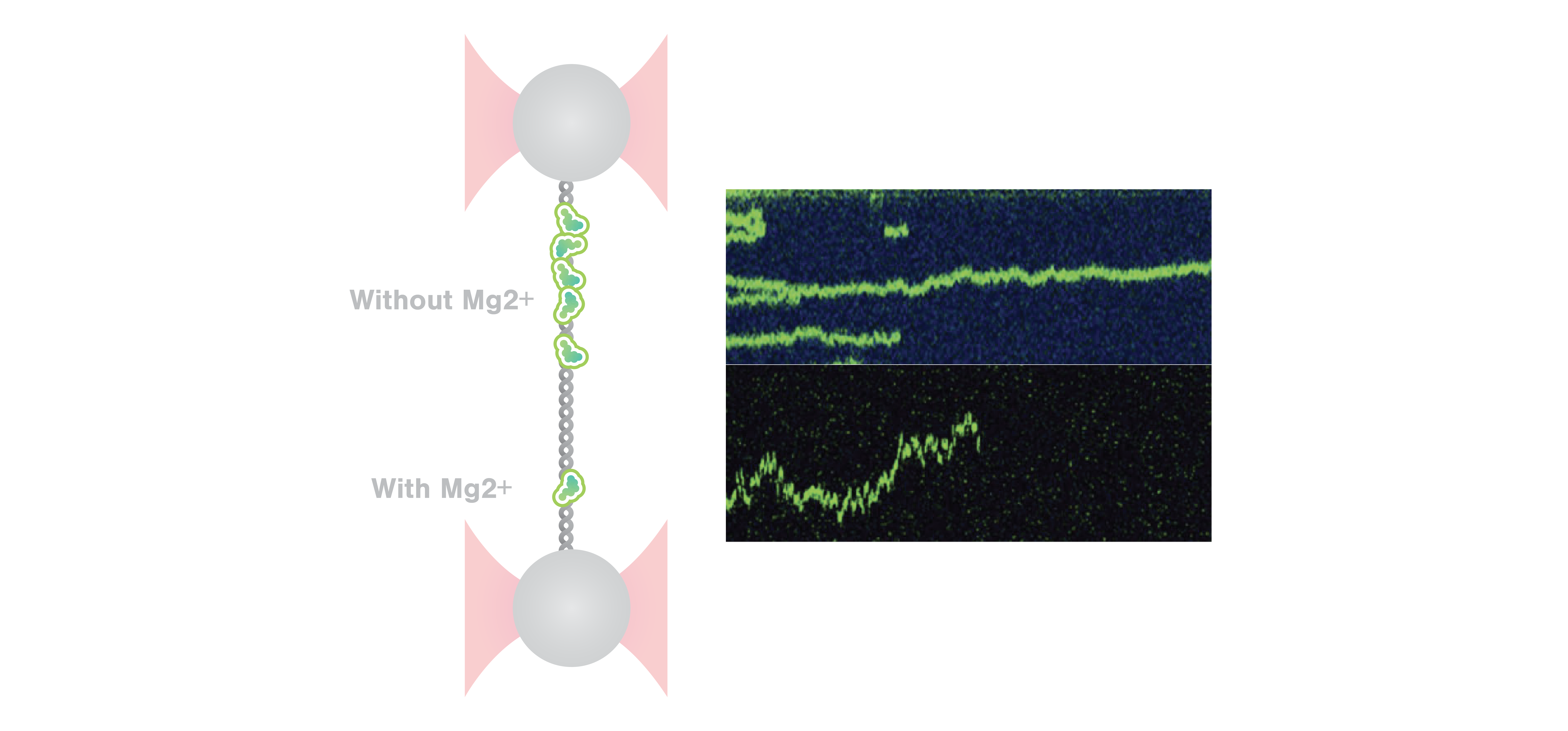
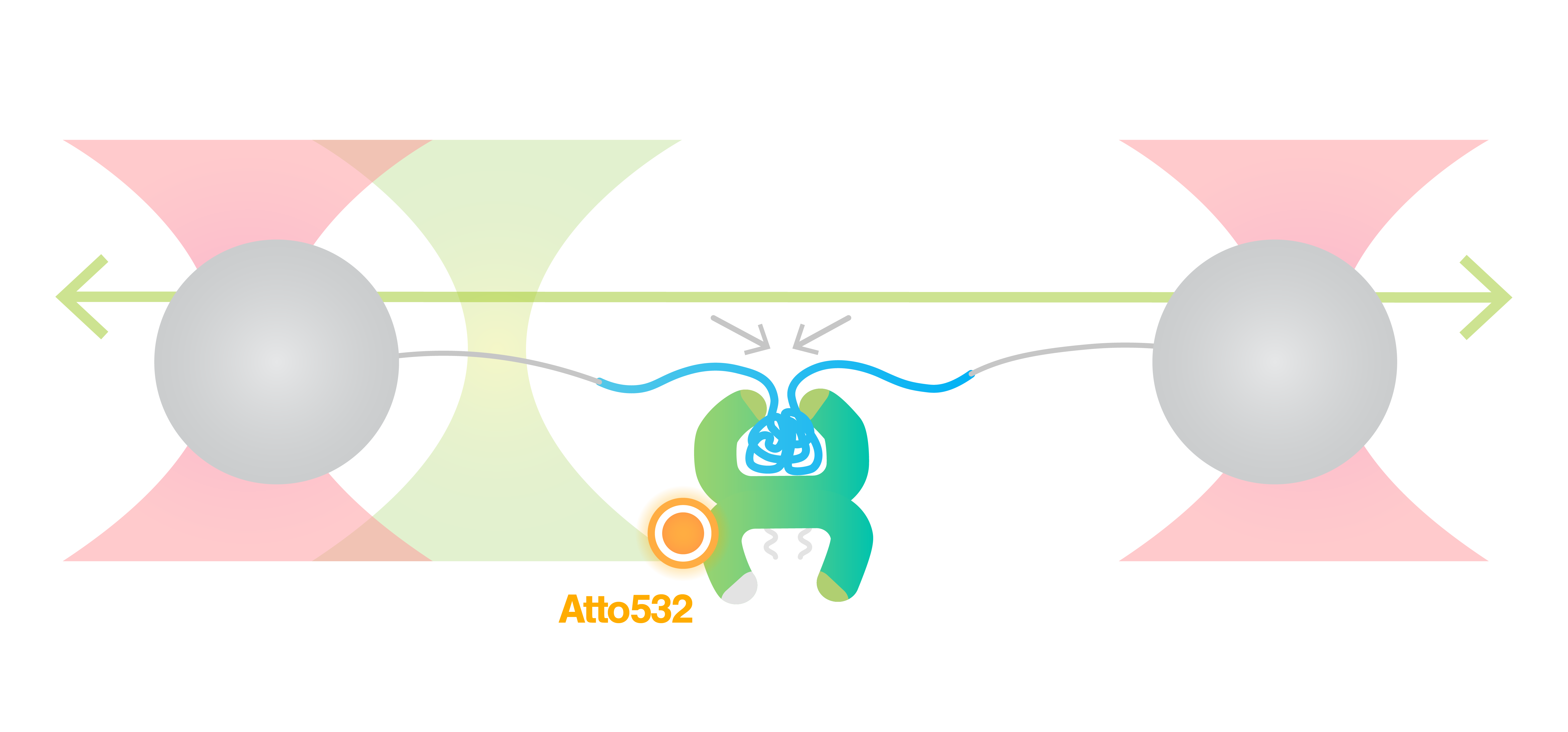
Using C-Trap® to unravel different binding modes of architectural DNA-binding proteins at different ionic conditions and DNA conformation - LUMICKS

Proteintech ChromoTek RFP-Trap™ Magnetic Agarose 10 Reactions Proteintech ChromoTek RFP-Trap™ Magnetic Agarose | Fisher Scientific

Comparison of protein characterization using In solution and S-Trap digestion methods for proteomics - ScienceDirect